News
The websites of Gao Lab have migrated into the cloud. Users can find the new url below.

http://cope.gao-lab.org/
Cope: accurately annotate compound effects of genetic variants using a context-sensitive framework.

http://annolnc.gao-lab.org/
AnnoLnc2: the one-stop portal to systematically annotate novel lncRNAs for human and mouse.

http://gsds.gao-lab.org/
GSDS 2.0: an upgraded gene feature visualization server.
http://atrm.gao-lab.org/
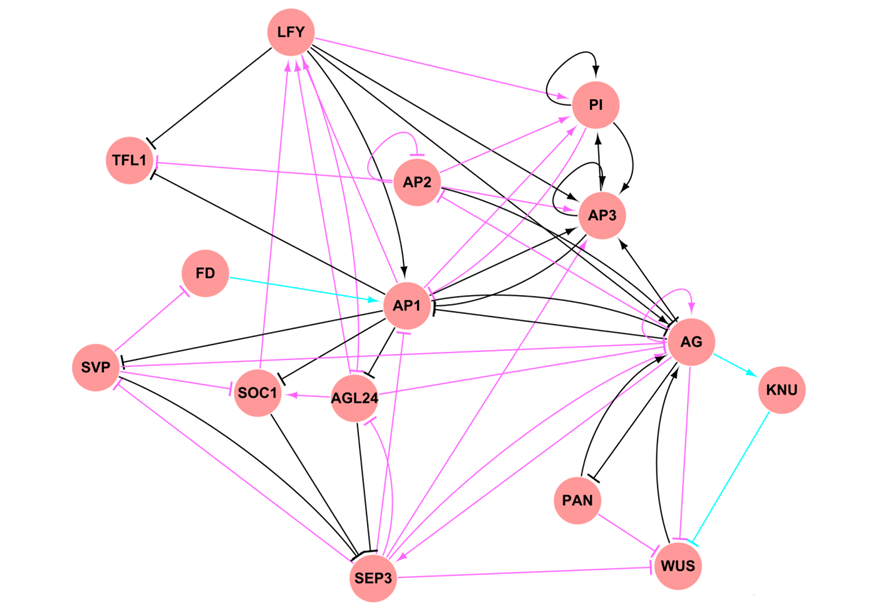
ATRM: arabidopsis transcriptional regulatory map.

http://planttfdb.gao-lab.org/
PlantTFDB: plant transcription factor database.

http://plantregmap.gao-lab.org/
PlantRegMap: plant transcriptional regulatory map.

http://cblast.gao-lab.org/
Cell BLAST: Searching large-scale scRNA-seq databases via unbiased cell embedding.

http://reflnc.gao-lab.org/
RefLnc: An expanded landscape of human long non-coding RNAs.

http://reva.gao-lab.org/
REVA: A Well-curated Database for Human Expression-modulating Variants.

http://carmen.gao-lab.org/
CARMEN: Computational Assessment of the Regulation-Modulating Potential for Noncoding Variants.

http://weblab.gao-lab.org/
WebLab: a data-centric, knowledge-sharing bioinformatic platform.

http://abrowse.gao-lab.org/
ABrowse: a customizable next-generation genome browser framework.

http://cpc.gao-lab.org/
CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine.

http://cpc2.gao-lab.org/